
Automated Parametrization of Biomolecular Force Fields from Quantum Mechanics/Molecular Mechanics (QM/MM) Simulations through Force Matching | Journal of Chemical Theory and Computation

Molecular Dynamics Simulations with Quantum Mechanics/Molecular Mechanics and Adaptive Neural Networks | Journal of Chemical Theory and Computation

Frontiers | Unnatural Amino Acid: 4-Aminopyrazolonyl Amino Acid Comprising Tri-Peptides Forms Organogel With Co-Solvent (EtOAc:Hexane)
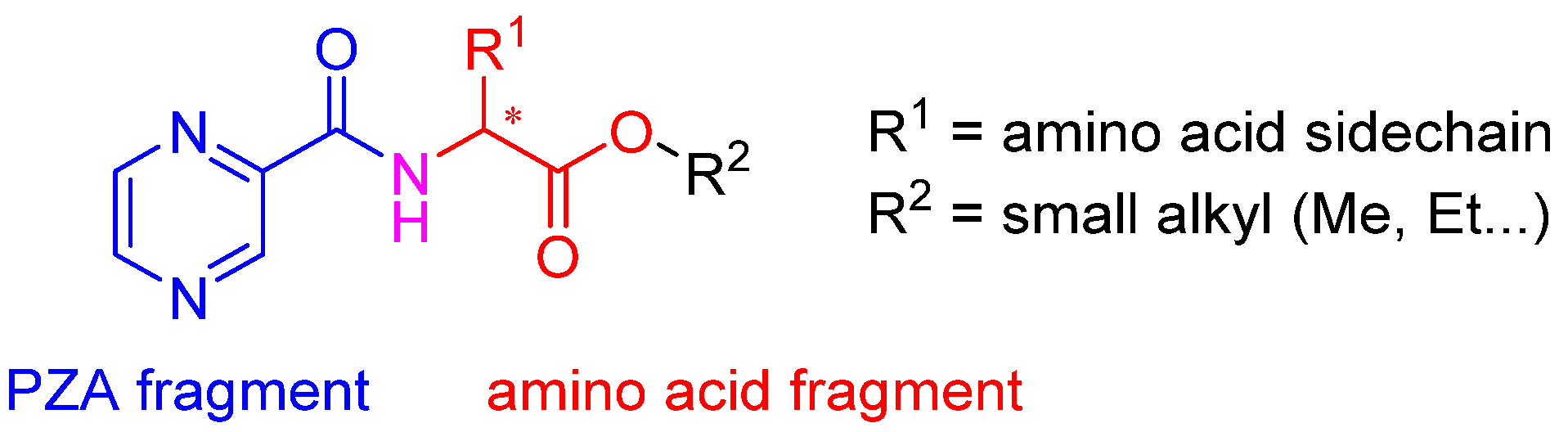
Molecules | Free Full-Text | N-Pyrazinoyl Substituted Amino Acids as Potential Antimycobacterial Agents—the Synthesis and Biological Evaluation of Enantiomers

Molecules | Free Full-Text | N-Pyrazinoyl Substituted Amino Acids as Potential Antimycobacterial Agents—the Synthesis and Biological Evaluation of Enantiomers
![PDF] Prediction of Mycobacterium tuberculosis pyrazinamidase function based on structural stability, physicochemical and geometrical descriptors | Semantic Scholar PDF] Prediction of Mycobacterium tuberculosis pyrazinamidase function based on structural stability, physicochemical and geometrical descriptors | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/89e829d38a85a7031554fe8812c46416b388e3ea/11-Table2-1.png)
PDF] Prediction of Mycobacterium tuberculosis pyrazinamidase function based on structural stability, physicochemical and geometrical descriptors | Semantic Scholar

Molecular Dynamics Simulations with Quantum Mechanics/Molecular Mechanics and Adaptive Neural Networks | Journal of Chemical Theory and Computation

X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease | Science

Pyrazinamide Susceptibility Is Driven by Activation of the SigE-Dependent Cell Envelope Stress Response in Mycobacterium tuberculosis | mBio






